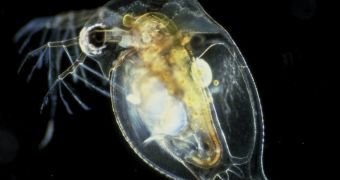
For a long time, researchers have been trying to identify the animal that features the most genes in its genetic code. It was at first believed that humans ranked first, but that turned out not to be true. In a new study, experts determined that the near-microscopic freshwater crustacean Daphnia pulex (water flea) is the winner of this “competition.”
Researchers have determined some time ago that the human genome featured around 23,000 genes, which seemed a lot at the time. However, the water flea puts us to shame with its 31,000 genes.
D. pulex is only the first crustacean to have its genome sequenced in its entirety, so subsequent studies might determine that other, similar creatures have even more genes. Researchers published details of the water flea genome in the latest issue of the top journal Science,
An international network of scientists was behind the new work. The collaboration is called the Daphnia Genomics Consortium, and it is led by experts with the US Department of Energy's (DOE) Joint Genome Institute and Indiana University (IU) Center for Genomics and Bioinformatics (CGB).
The investigation was funded by the Division of Environmental Biology (DEB) at the US National Science Foundation (NSF). The analysis is bound to inspire other investigators in carrying our similar studies on less known species.
“Daphnia's high gene number is largely because its genes are multiplying, creating copies at a higher rate than other species. We estimate a rate that is three times greater than those of other invertebrates and 30 percent greater than that of humans,” says John Colbourne.
The investigator is the lead researcher on the project, and also the genomics director at the CGB.
“This analysis of the Daphnia genome significantly advances our understanding of how an organism's genome interacts with its environment both to influence genome structure and to confer ecological and evolutionary success,” adds Saran Twombly, who is a DEB program director.
“This gene-environment interplay has, to date, been studied in model organisms under artificial, laboratory conditions,” the NSF official goes on to explain.
“Because the ecology of Daphnia pulex is well-known, and the organism occurs abundantly in the wild, this analysis provides unprecedented insights into the feedback between genes and environment in a real and ever-changing environment,” Twombly explains.
In addition to the NSF, the main supporters of the new study were the DOE, Lilly Endowment Inc., Roche NimbleGen Inc., the US National Institutes of Health (NIH), the Department of Health and Human Services (DHHS), and the Indiana University.
 14 DAY TRIAL //
14 DAY TRIAL //