A collaboration of investigators from the Massachusetts Institute of Technology (MIT) and the Harvard University announces the development of a new technology that enables them to rewrite the genetic code of a living cell with great proficiency and speed.
This approach enables researchers to create large-scale edits of the cellular genome, which means that they could tweak an organisms while it is still alive. What this means is that researchers could soon become capable of building custom cells.
Some of these cells could be used to construct molecules that cannot be found in nature, such as all-new proteins and enzymes, among others. At the same time, they could also learn how to produce microorganisms that are resistant to viral infections.
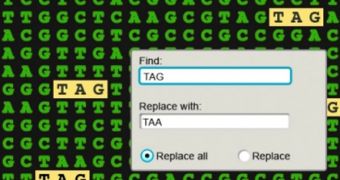
The way the research team explains the process is by way of analogy with the find-and-replace function that is standard to all programs dealing with word processing. With the new technique, experts simply input the chain of nucleotides that they want switched, and watch as this happens across the genome.
In lab experiments conducted to test the method, the joint team was able to edit the genome of the Escherichia coli bacteria in hundreds of places, without triggering any type of discernible malfunction in the way these unicellular organisms generally function.
Details of the research effort were published in the July 15 issue of the top journal Science. “We did get some skepticism from biologists early on,” investigator Peter Carr says of this research effort.
“When you’re making so many intentional changes to the genome, you might think something’s got to go wrong with that,” adds the expert, who is a MIT Lincoln Laboratory senior research staff member.
The new method is the result of a 7-year collaboration between Harvard Medical School professor of genetics George Church and MIT Media Lab associate professor Joseph Jacobson.
Together with a number of other experts from the two institutions, the experts combined a technology called multiplex automated genome engineering (MAGE) with another one dubbed conjugative assembly genome engineering (CAGE).
By using the two technologies together, it could become possible to engineer bacteria for industrial applications. These microorganisms would be more efficient at producing whatever chemical is expected at them, but would also be unable to spread their genetic material into natural environments.
 14 DAY TRIAL //
14 DAY TRIAL //