Researchers at the Georgia Institute of Technology (GIT) have developed a novel technique of peering inside living cells, which allows them to visualize single molecules of ribonucleic acid (RNA), with much more ease than existing methods. With the use of the new instrument, experts could gain even more access into the daily “operations” going on inside living cells, as opposed to just being stuck with investigating dead ones.
With existing imaging techniques, researchers have to place a large amount of fluorescent molecules inside the cells to evidence the RNA molecules, or even to inject a large number of synthetic RNA into the organism. But the GIT team has developed a new approach to viewing the acid, by creating a type of fluorescent probe that moves past these problems. A scientific paper detailing the accomplishment is published in the April 6th issue of the journal Nature Methods.
“The probes we designed shine bright, are small and easy to assemble, bind rapidly to their targets, and can be imaged for hours. These characteristics make them a great choice for studying the movement and location of RNA inside a single cell and the interaction between RNA and binding proteins,” Georgia Tech and Emory University Wallace H. Coulter Department of Biomedical Engineering Assistant Professor Philip Santangelo explains.
“The great thing about these probes is that they recognize RNA sequences and bind to them using the same base pairing most people are familiar with in regards to DNA. By adding only a few probes that would bind to a region of RNA, we gained the ability to distinguish a targeted RNA molecule from a single unbound probe because the former lit up two or three times brighter,” the scientist adds.
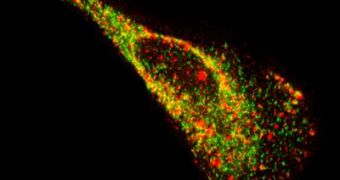
The new probes are made up of fluorophores – small fluorescent molecules – attached to modified nucleic acid sequences and a specific protein. The researchers have been amazed to notice that the mixture has provided single-molecule sensitivity, and to learn that the effects of the fluorescence can last up to several hours. The probes can attach to both native RNA and viral, non-engineered RNA in the target cells.
“We observed substantial transient interactions between proteins and viral RNA molecules that I don't think had ever been seen before with non-engineered RNA. We saw one of the proteins move into a viral RNA granule and reside within it for over a minute before it was released, and we also saw another protein that appeared to dock with a viral RNA granule,” Santangelo says.
“We are excited to use this imaging strategy to study how single viral RNAs travel from the nucleus of a cell to a virus assembly site, how mRNAs are regulated by location and time, and RNA trafficking in neurons,” he concludes.
 14 DAY TRIAL //
14 DAY TRIAL //