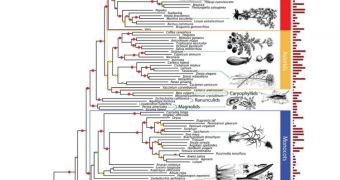
The evolutionary course of 150 plant species has been charted with extreme precision in a new scientific investigation, announce researchers at the Cold Spring Harbor Laboratory. This genome-based tree of life is the largest of its type ever constructed.
The CSHL team worked together with colleagues at the New York Botanical Garden, the New York University and the American Museum of National History. The work was published in the December 16 issue of the journal PLoS Genetics, which is edited by the Public Library of Science.
In this investigation, researchers constructed the tree of life using advanced genome-wide analysis techniques, which revealed and related gene structures and functions to each other. This allowed the team to construct a map of the relationships between the 150 species.
This new type of approach to conducting genetic analyses is called functional phylogenomics, and it allows researchers to figure out how species evolved from common ancestors over long stretches of time. But the work also has practical applications.
Using the new dataset, it may be possible to manipulate seeds genetically so that they become more productive and safe. This would lead to an increase in agricultural production, but also to an improvement in the impact crops have on the environment.
“Ever since Darwin first described the ‘abominable mystery’ behind the rapid explosion of flowering plants in the fossil record, evolutionary biologists have been trying to understand the genetic and genomic basis of the astounding diversity of plant species,” researcher Rob DeSalle explains.
“Having the architecture of this plant tree of life allows us to start to decipher some of the interesting aspects of evolutionary innovations that have occurred in this group,” he goes on to say.
The expert holds an appointment as a curator at the AMNH Division of Invertebrate Zoology, and at the Sackler Institute for Comparative Genomics. He was also a corresponding author on the new paper.
Funding for the research was provided through the US National Science Foundation (NSF) Plant Genome Program, to the New York Plant Genomics Consortium. The species the group analyzed belonged to all major seed plant groups.
The CSHL supercomputers had to process nearly 23,000 sets of genes, and order them so that their evolutionary links become obvious. This was only made possible by new algorithms created specifically for this investigation.
“In our novel approach, we create the phylogeny based on all the genes in a genome, and then use the phylogeny to identify which genes provide positive support for the divergence of species,” NYU Center for Genomics and Systems Biology Gloria Coruzzi says. She was the principal investigator on the study.
 14 DAY TRIAL //
14 DAY TRIAL //