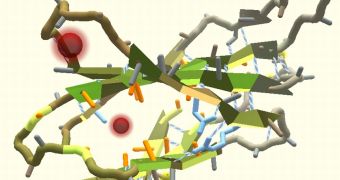
A new research paper has shown that players engaged in the online video game Foldit have managed to determine, through crowdsourcing, the optimal structure of a very important protein that could be one of the main elements needed to develop new drugs that can reduce and even eliminate AIDS from the body of a patient.
The report comes from the University of Washington and states that players of Foldit have managed to create a model for a protein found in the Mason-Pfizer monkey virus, which is responsible for a simian form of AIDS.
The research shows that the player created protein model was more accurate than that created by supercomputers after years of simulations.
Foldit allows players to work in teams in order to create the best scoring model of a protein within certain rules.
The research paper says, “The critical role of Foldit players in the solution of the M-PMV PR structure shows the power of online games to channel human intuition and three-dimensional pattern-matching skills to solve challenging scientific problems.”
It adds, “Although much attention has recently been given to the potential of crowdsourcing and game playing, this is the first instance that we are aware of in which online gamers solved a longstanding scientific problem.”
The research paper says that human players were not always better than computers at creating better performing protein models but that the competition model encouraged them to make significant and risky changes to their models, leading to a number of improvements that would otherwise not have been noticed.
Until now, researchers have concentrated on using spare computing power from gaming rigs to support research, through initiatives like Folding@Home or Progress through Processors but it seems that making the entire concept competitive and directly involving gamers might be a better idea in the long term.
 14 DAY TRIAL //
14 DAY TRIAL //