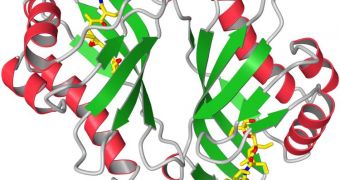
An international collaboration of researchers was recently able to create a model of the unique enzyme Lsd19, which researchers say common species of soil bacteria use to create compounds with natural antibiotic properties.
Ever since microorganisms were found to be able to create antibiotics, chemists have been wondering how these lifeforms perform the necessary reactions to reach those compounds. The reactions form six-membered cyclic ether rings, a process that was demonstrated to be kinetically-unfavorable.
Studies then focused on figuring out how bacteria such as Streptomyces lasaliensis produced these chemicals. Analyses conducted over the years revealed that the enzyme Lsd19 plays a critical role in underlying these reactions.
But the structure of the protein has never been fully understood until now. Researchers in the United States, the United Kingdom, Japan and Singapore used the Stanford Synchrotron Radiation Lightsource (SSRL), at the SLAC National Accelerator Laboratory, for this research.
The advanced light source enabled scientists to use an imaging technique called X-ray crystallography on the enzyme. The approach involves freezing the target molecules inside crystals, and then bombarding those crystals with intense X-ray blasts.
After conducting the experiment a sufficient number of times, scientists can create a 3D model of the molecules being researched. This is precisely what the team did for Lsd19. Investigators say that this finding may contribute extensively to developing new ways of synthesizing polyether drugs.
The work was led by National University of Singapore (NUS) Faculty of Science assistant professor Chy-Young Kim. Details of their research were published in the March 5 issue of the top scientific journal Nature.
“Our study has broad implications because the six-membered cyclic ether is a common structural feature found in hundreds of drug molecules produced by nature,” Dr. Kim explains.
“We have analysed the genes of six other organisms that produce similar polyether drugs and we are now confident that the biosynthetic strategy we have uncovered is also used by those organisms,” the principal investigator adds .
“The bugs have taught us a valuable chemistry lesson. With a new understanding of how nature synthesises the six-membered rings, chemists may be able to develop new methods to produce polyether drugs with ease in the laboratory,” the researcher says, quoted by AlphaGalileo.
 14 DAY TRIAL //
14 DAY TRIAL //