Scientists in the United States announce that they are now able to produce computer simulations of interactions taking place inside in vivo biological systems. Thus far, this has only been possible in isolated systems, on Petri dishes.
Some of the most common biological systems include cells, tissues and organs. All these arrangements are incredibly complex, and yet they only function properly when in harmony with others.
This is why analyzing their function in detail is such a complex task. Keeping track on the billions of cells making up a single organ is a monumental challenge, and one that cannot be easily met.
[MIT=1]Biologists have lately resorted to computer models to try and figure out what's happening inside even the most basic cells. This approach is called systems biology, and it is apparently working.
The thing that makes regular studies impossible is the sheer number of molecules that interacts within a brief period of time. Literally thousands of proteins influence just as many processes any given second.
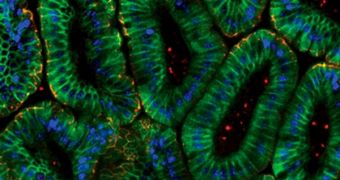
Until recently, computational biology has only been used to model the behavior of cells grown inside Petri dishes in scientific labs. What the new study did was demonstrate it's possible to use it for modeling the interactions taking place inside living biological systems as well.
This was accomplished by researchers at the Massachusetts Institute of Technology (MIT) and the Massachusetts General Hospital (MGH). The joint team developed a brand new computational model.
The simulation provides unique insight into how intestinal cells in mice act in the presence of a natural chemical called tumor necrosis factor (TNF). Details of the achievement are published in the March 22 online issue of the esteemed medical journal Science Signaling.
“You're not likely to explain most diseases in terms of one genetic deficit or one molecular impairment,” explains the head of the MIT Department of Biological Engineering, Douglas Lauffenburger.
“You need to understand how many molecular components, working in concert, give rise to how cells and tissues are formed – either properly or improperly,” adds the expert, also the senior author of the new research paper.
He adds that the new models could perhaps be used to model the complexity of cancer and its tumors, as well as the onset, development and manifestations of other complex diseases plaguing the world.
“We expect that our ability to predict which targets, which drugs and which patients to bring together in the context of cancer treatment will require a deeper understanding of the complex signaling pathways that exist in cancer. This approach will help us get there,” says Tyler Jacks.
He is the director of the David H. Koch Institute for Integrative Cancer Research at MIT,
 14 DAY TRIAL //
14 DAY TRIAL //